Publication: An R package for phenotyping health-related outcomes in the UK Biobank
Ming Wai Yeung and Niek Verweij developed an R package (ukbpheno) that can efficiently process UK Biobank phenotypic data. The protocol is published in STAR protocols and detailed explanation with example analyses have been provided.
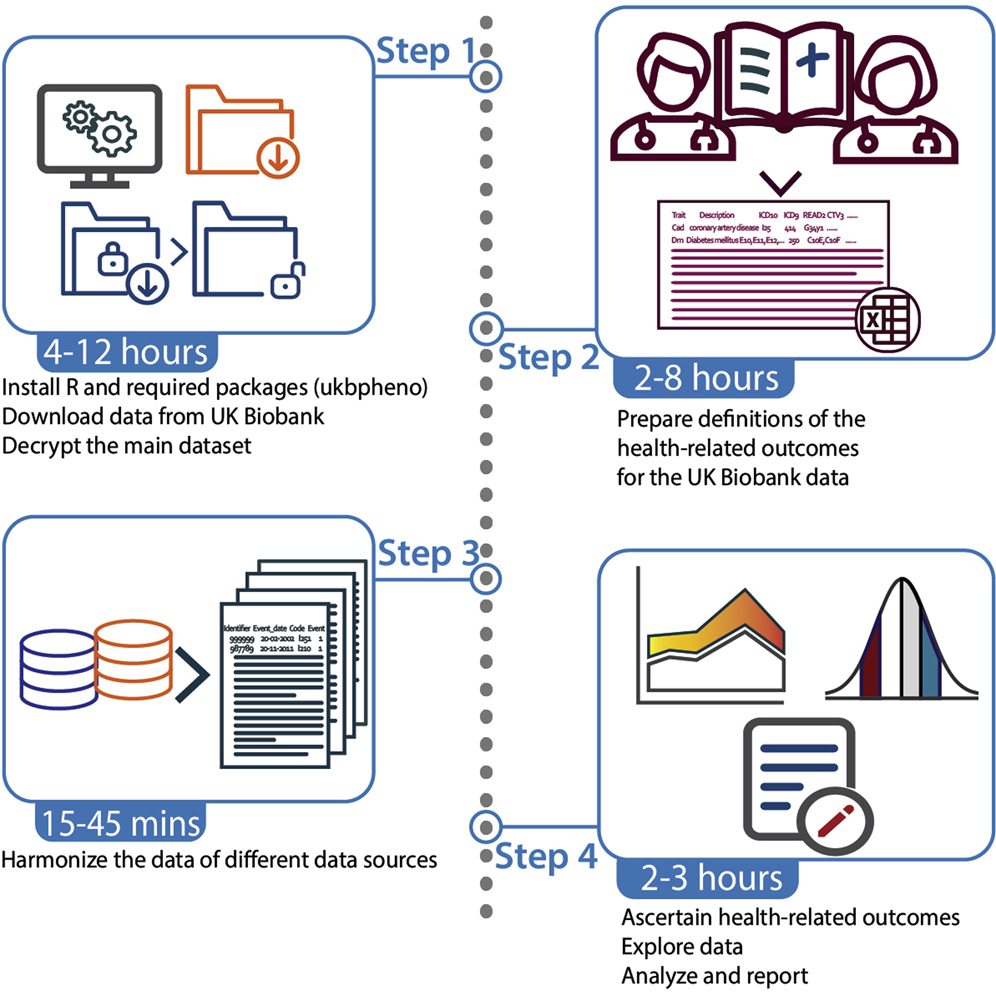
The UK Biobank is one of the largest open-access resource for health research worldwide. While it is an invaluable resource, the magnitude of data volume, complexity of information collected as well as the longitudinal nature of data may pose a challenge for researchers to analyze health related outcomes.
To this end, Ming Wai Yeung and Niek Verweij developed an R package ukbpheno to harmonise and create longitudinal phenotypes in UK Biobank to aid explore and analysis of the dataset in an efficient manner. Additionally, the authors wrote a protocol following an example outcome (diabetes) to illustrate the usage of ukbpheno from access of the data to sample analyses in a step-by-step manner. ukbpheno is open source and freely available to the research community.
This package can efficiently process UK Biobank phenotypic data. It helps to provide reproducible and semi-automatic generation of binary health-related phenotypes. It has also visualization tools to aid data exploration and phenotype definition. The authors also provided example analyses for common among genetic epidemiological studies.
To check the protocol, please click here.
To check the R package, please click here.